February 2023
Abstract
Objective
To examine the oral microbiome in the context of oral cavity squamous cell carcinoma.
Study Design
Basic science research. Setting Academic medical center.
Methods
Oral swabs were collected from patients presenting to the operating room for management of oral cavity squamous cell carcinoma and from age- and sex-matched control patients receiving surgery for unrelated benign conditions. 16S ribosomal RNA (rRNA) sequencing was performed on genetic material obtained from swabs. A bacterial rRNA gene library was created and sequence reads were sorted into taxonomic units.
Results
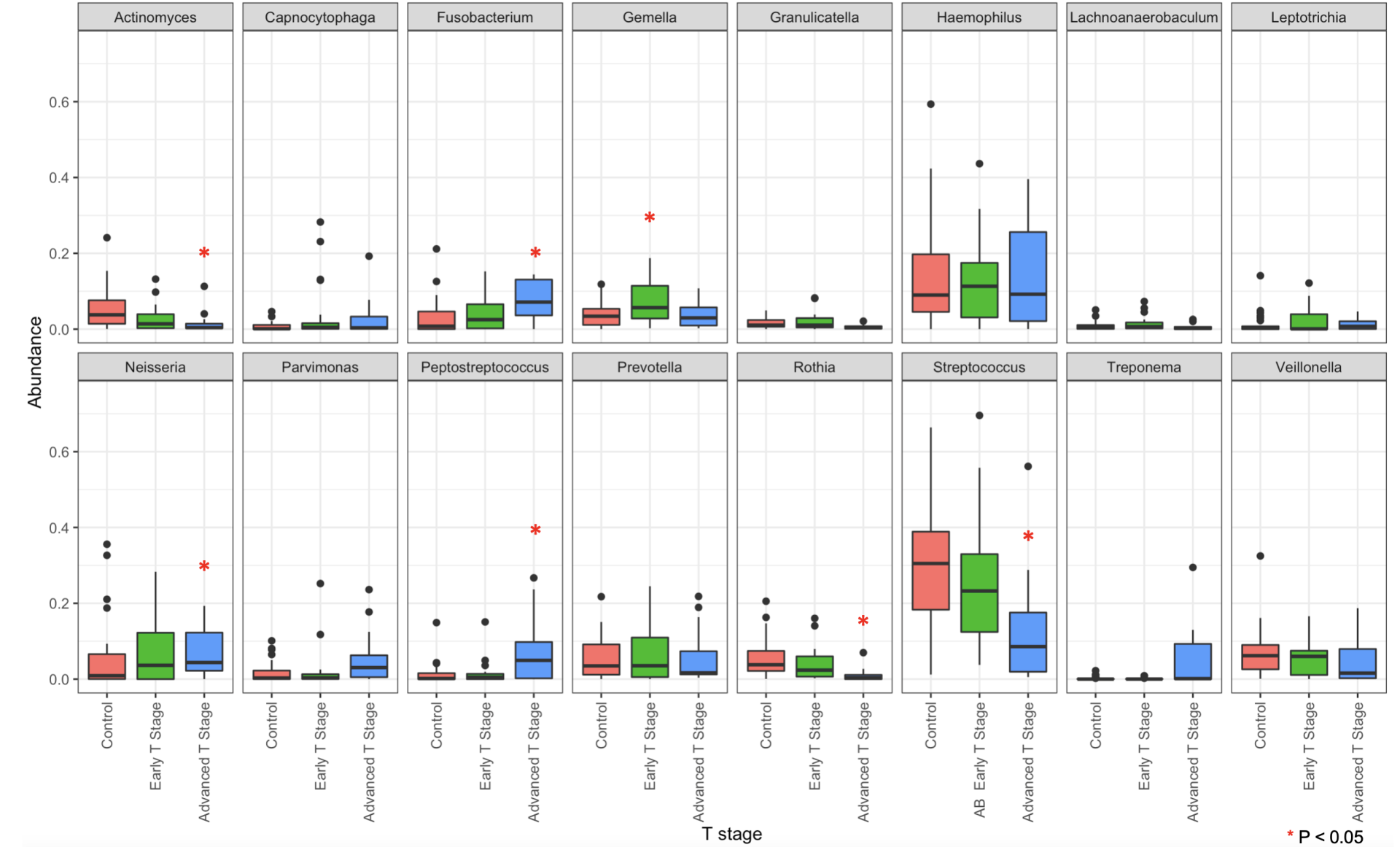
Thirty-one control patients (17 males) and 35 cancer patients (21 males) were enrolled. Ages ranged from 23 to 89 (median 63) for control patients and 35 to 86 (median 66) for cancer patients. Sixty-one percent of control patients and 63\% of cancer patients were smokers. 16S analyses demonstrated a significant decrease in Streptococcus genera in oral cancer patients (34.11\% vs 21.74\% of the population, p = .04). Increases in Fusobacterium, Peptostreptococcus, Parvimonas, and Neisseria were also found. The abundance of these bacteria correlated with tumor T-stage.
Conclusion
16S rRNA sequencing demonstrated changes in bacterial populations in oral cavity cancer and its progression compared to noncancer controls. We found increases in bacteria genera that correspond with tumor stage—Fusobacteria, Peptostreptococcus, Parvimonas, Neisseria, and Treponema. These data suggest that oral cancer creates an environment to facilitate foreign bacterial growth, rather than implicating a specific bacterial species in carcinogenesis. These bacteria can be employed as a potential marker for tumor progression or interrogated to better characterize the tumor microenvironment.
